真固生物 学术海报
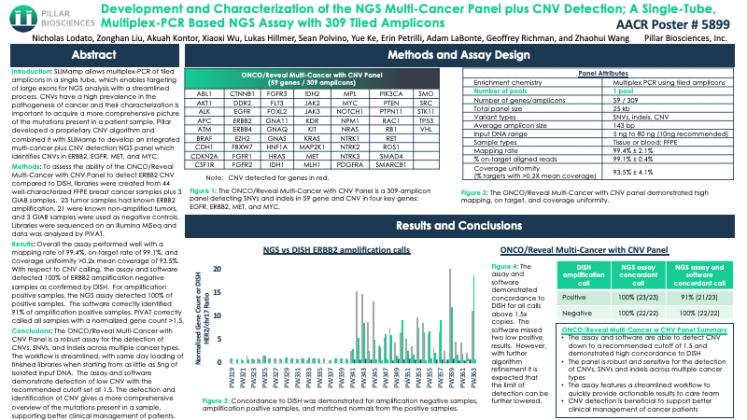
Development and Characterization of the NGS Multi-Cancer Panel plus CNV Detection; A Single-Tube, Multiplex-PCR Based NGS Assay with 309 Tiled Amplicons
- Nicholas Lodato, Zonghan Liu, Akuah Kontor, Xiaoxi Wu, Lukas Hillmer, Sean Polvino, Yue Ke, Erin Petrilli, Adam LaBonte, Geoffrey Richman, and Zhaohui Wang
- AACR 2020
-
SLIMamp allows multiplex-PCR of tiled amplicons in a single tube, which enables targeting of large exons for NGS analysis with a streamlined process. CNVs have a high prevalence in the pathogenesis of cancer and their characterization is important to acquire a more comprehensive picture of the mutations present in a patient sample. Pillar developed a proprietary CNV algorithm and combined it with SLIMamp to develop an integrated multi-cancer plus CNV detection NGS panel which identifies CNVs in ERBB2, EGFR, MET, and MYC.
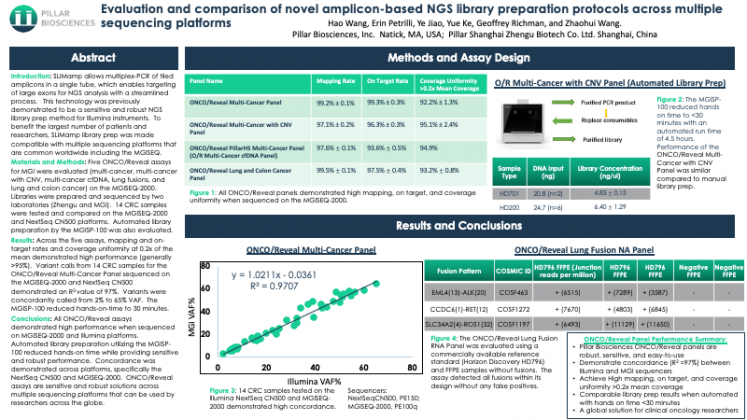
Evaluation and comparison of novel amplicon-based NGS library preparation protocols across multiple sequencing platforms
- Hao Wang, Erin Petrilli, Ye Jiao, Yue Ke, Geoffrey Richman, and Zhaohui Wang. Pillar Biosciences, Inc. Natick, MA, USA; Pillar Shanghai Zhengu Biotech Co. Ltd. Shanghai, China
- ESHG 2020
-
SLIMamp allows multiplex-PCR of tiled amplicons in a single tube, which enables targeting of large exons for NGS analysis with a streamlined process. This technology was previously demonstrated to be a sensitive and robust NGS library prep method for Illumina instruments. To benefit the largest number of patients and researchers, SLIMamp library prep was made compatible with multiple sequencing platforms that are common worldwide including the MGISEQ.
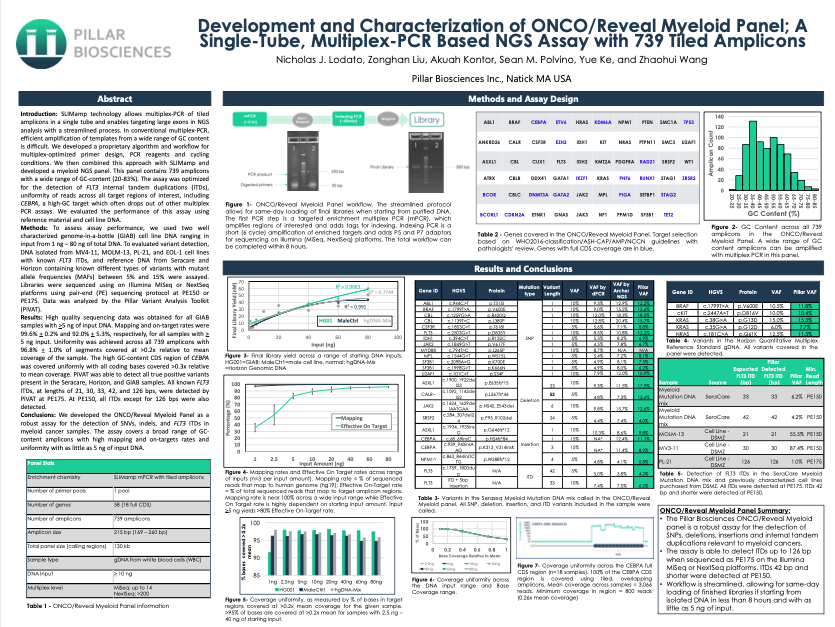
- Nicholas J. Lodato, Zonghan Liu, Akuah Kontor, Sean M. Polvino, Yue Ke, and Zhaohui Wang
- AMP 2019
-
SLIMamp technology allows multiplex-PCR of tiled amplicons in a single tube and enables targeting large exons in NGS analysis with a streamlined process. In conventional multiplex-PCR, efficient amplification of templates from a wide range of GC content is difficult. We developed a proprietary algorithm and workflow for multiplex-optimized primer design, PCR reagents and cycling conditions. We then combined this approach with SLIMamp and developed a myeloid NGS panel. This panel contains 739 amplicons with a wide range of GC-content (20-83%). The assay was optimized for the detection of FLT3 internal tandem duplications (ITDs), uniformity of reads across all target regions of interest, including CEBPA, a high-GC target which often drops out of other multiplex PCR assays. We evaluated the performance of this assay using reference material and cell line DNA.
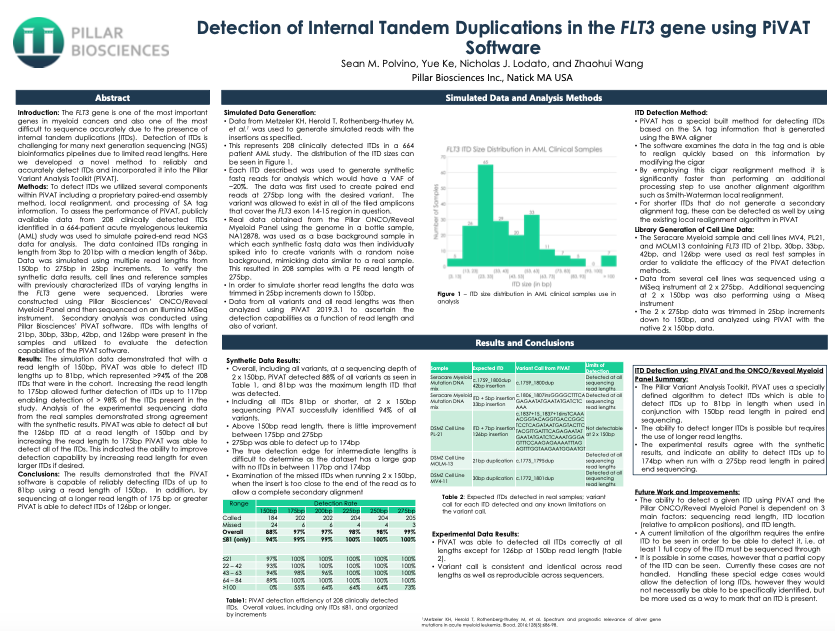
- Sean M. Polvino, Yue Ke, Nicholas J. Lodato, and Zhaohui Wang
- AMP 2019
-
The FLT3 gene is one of the most important genes in myeloid cancers and also one of the most difficult to sequence accurately due to the presence of internal tandem duplications (ITDs). Detection of ITDs is challenging for many next generation sequencing (NGS) bioinformatics pipelines due to limited read lengths. Here we developed a novel method to reliably and accurately detect ITDs and incorporated it into the Pillar Variant Analysis Toolkit (PiVAT).
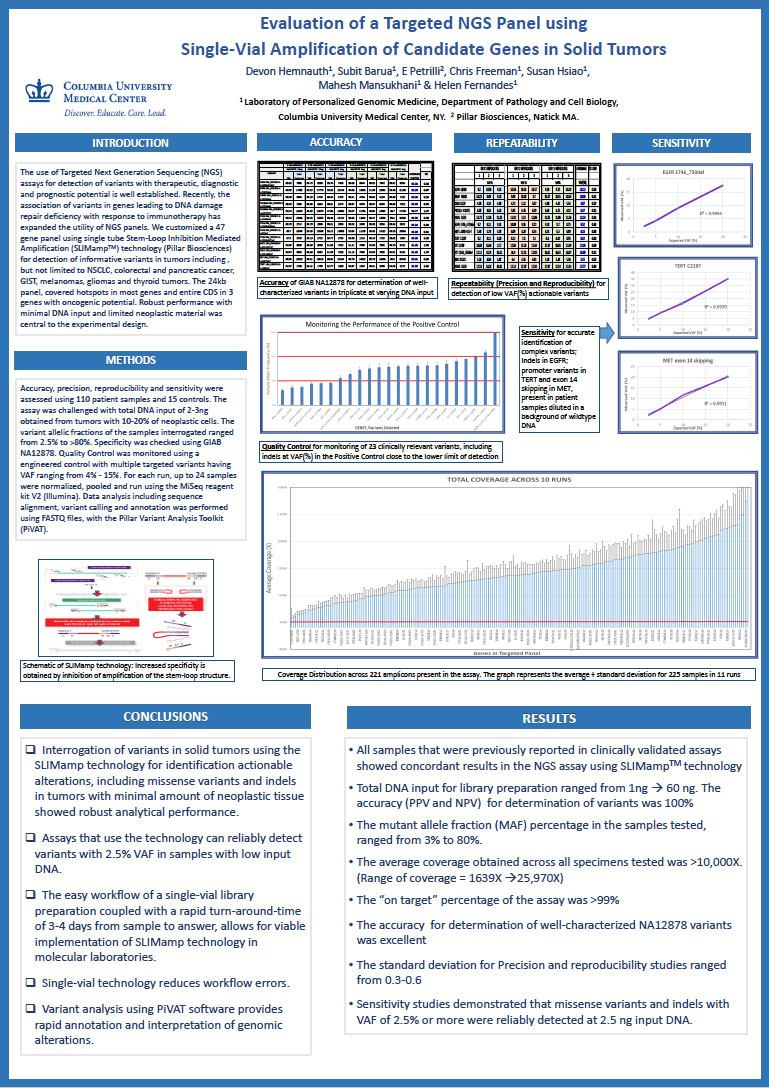
Evaluation of a Targeted NGS Panel using Single-Vial Amplification of Candidate Genes in Solid Tumors
- Devon Humnauth, Subit Barua, Erin Petrilli, Chris Freeman, Susan Hsiao, Mahesh Mansukhani, and Helen Fernandes
- AMP Global 2019
- The use of targeted Next Generation Sequencing (NGS) assays for detection of variants with therapeutic, diagnostic and prognostic potential is well established. Recently, the association of variants in genes leading to DNA damage repair deficiency with response to immunotherapy has expanded the utility of NGS panels. We customized a 47-gene panel using single-tube Stem-Loop Inhibition-Mediated Amplification (SLIMampTM) technology (Pillar Biosciences) for detection of informative variants in tumors including, but not limited to, NSCLC, colorectal and pancreatic cancer, GIST, melanomas, gliomas and thyroid tumors. The 24kb panel covered hotspots in most genes and entire CDS in 3 genes with oncogenic potential. Robust performance with minimal DNA input and limited neoplastic material was central to the experimental design.
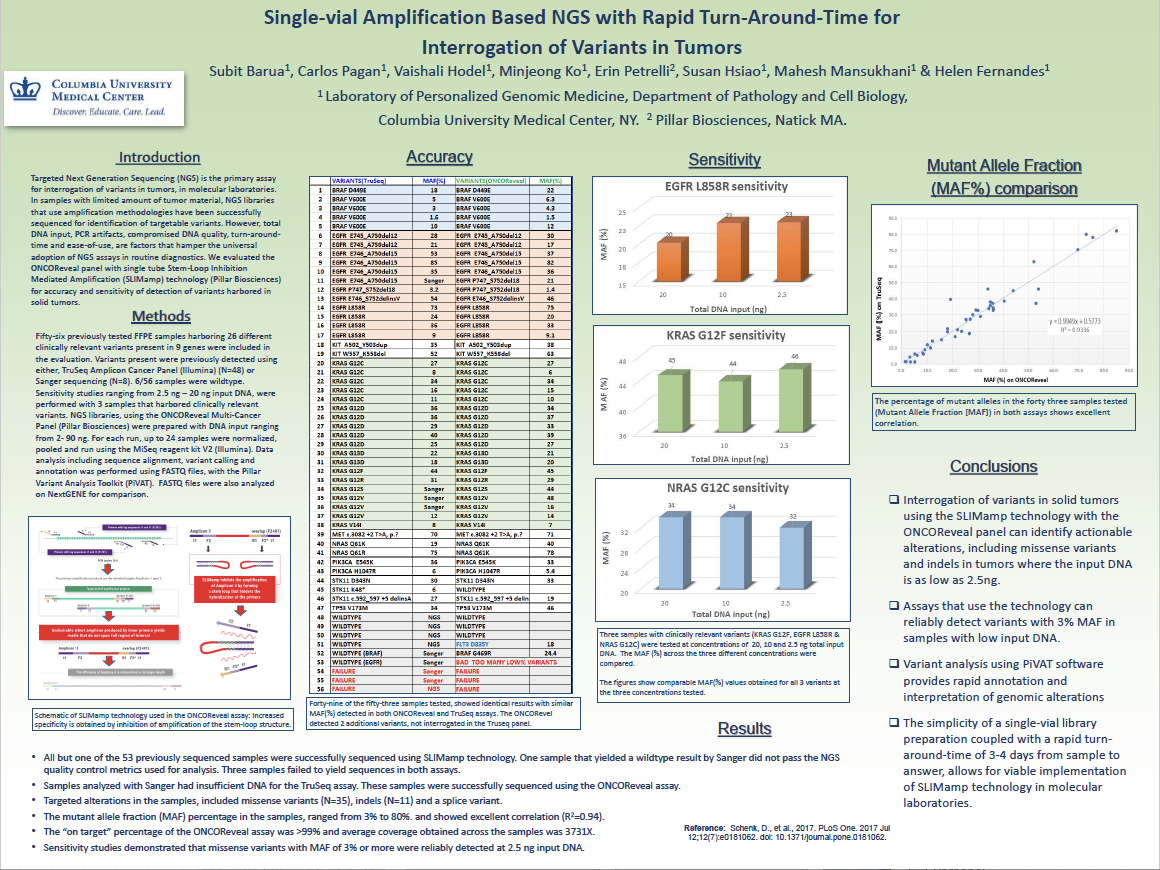
- Sabit Barua, Carlos Pagan, Vaishali Hodel, Minjeong Ko, Mahesh Mansukhani, Erin Petrelli, Susan Hsiao, & Helen Fernandes
- AMP 2018
- Fifty-six previously tested FFPE samples harboring 26 different clinically relevant variants present in 9 genes were included in the evaluation. The ONCO/Reveal panel identified actionable alterations, including missense variants and indels in tumors where the input DNA is as low as 2.5ng. The assay can reliably detect variants with 3% MAF in samples with low input DNA. Variant analysis using PiVAT software provides rapid annotation and interpretation of genomic alterations. The simplicity of a single-vial library preparation coupled with a rapid turn-around-time of 3-4 days from sample to answer, allows for viable implementation of SLIMamp technology in molecular laboratories.

- Martin Zillmann, Yue Ke, Nicholas Lodato, Melanie Prasol, Zhaohui Wang,
- AMP 2018
- We have demonstrated a rapid and robust primer design pipeline using the TP53 oncogene as a model system. The resulting primers were shown to provide uniform coverage across TP53 and sensitive variant detection at FFPE-derived DNA inputs greater than 1 ng. In addition, the platform has been shown to be extensible to very large primer pools.
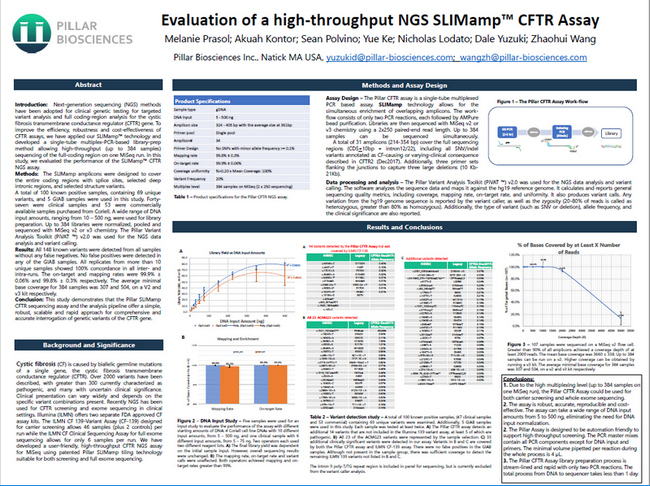
- Melanie Prasol, Akuah Kontor, Sean Polvino, Yue Ke, Nicholas Lodato, Dale Yuzuki, Zhaohui Wang
- AGBT 2018
- To improve the robustness, efficiency and cost-effectiveness of cystic fibrosis transmembrane conductance regulator (CFTR) gene assays, we have applied our SLIMamp assay technology to enable up to 384-plex sequencing of the full coding region on a single MiSeq instrument run. 148 variants were examined across 53 commercial samples with known variants, 47 clinical samples with known positive status and 5 Genome-in-a-Bottle (GIAB) samples.

Development and Evaluation of a Lung Cancer Fusion NGS Panel
- Melanie Prasol, Akuah Kontor, Sean Polvino, Yue Ke, Nicholas Lodato, Dale Yuzuki, Zhaohui Wang
- Next Gen Dx 2018
- This poster reviews the assay principle behind Pillar Biosciences’ Lung Cancer Fusion RNA Panel, the types of fusions interrogated, and presents results from 40 samples with known fusions from a variety of sources.
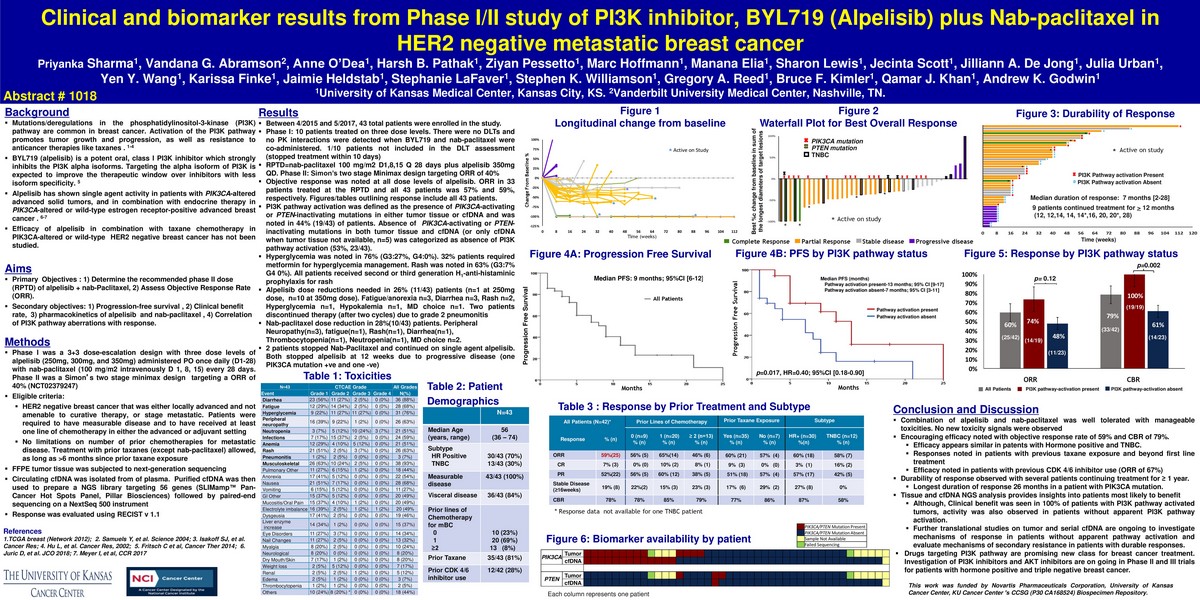
- Priyanka Sharma, Vandana G. Abramson, Anne O´Dea, Harsh B. Pathank, Ziyan Pessetto, Marc Hoffman, Marc Hoffmann, Manana Elia, Sharon Lewis, Jecinta Scott, Jilliann A. De Jong, Julia Urban, Yen Y. Wang, Karissa Finke, Jaimie Heldstab, Stephanie LaFaver, Stephen K. Williamson, Gregory A. Reed, Bruce F. Kimler, Qamar J. Khan, Andrew K. Godwin,
- ASCO 2018
- At the June 2018 American Society for Clinical Oncology conference held in Chicago IL (USA), the Kansas University Medical Center presented a poster describing the biomarker results of a Phase I-II clinical study of an experimental PI3K inhibitor called BYL719 (Apelisib) from Novartis. They used the ONCO/Reveal Multi-Cancer Panel from Pillar Biosciences on circulating cell-free DNA as part of their study. More information about this sensitive and efficient enrichment assay can be found here.
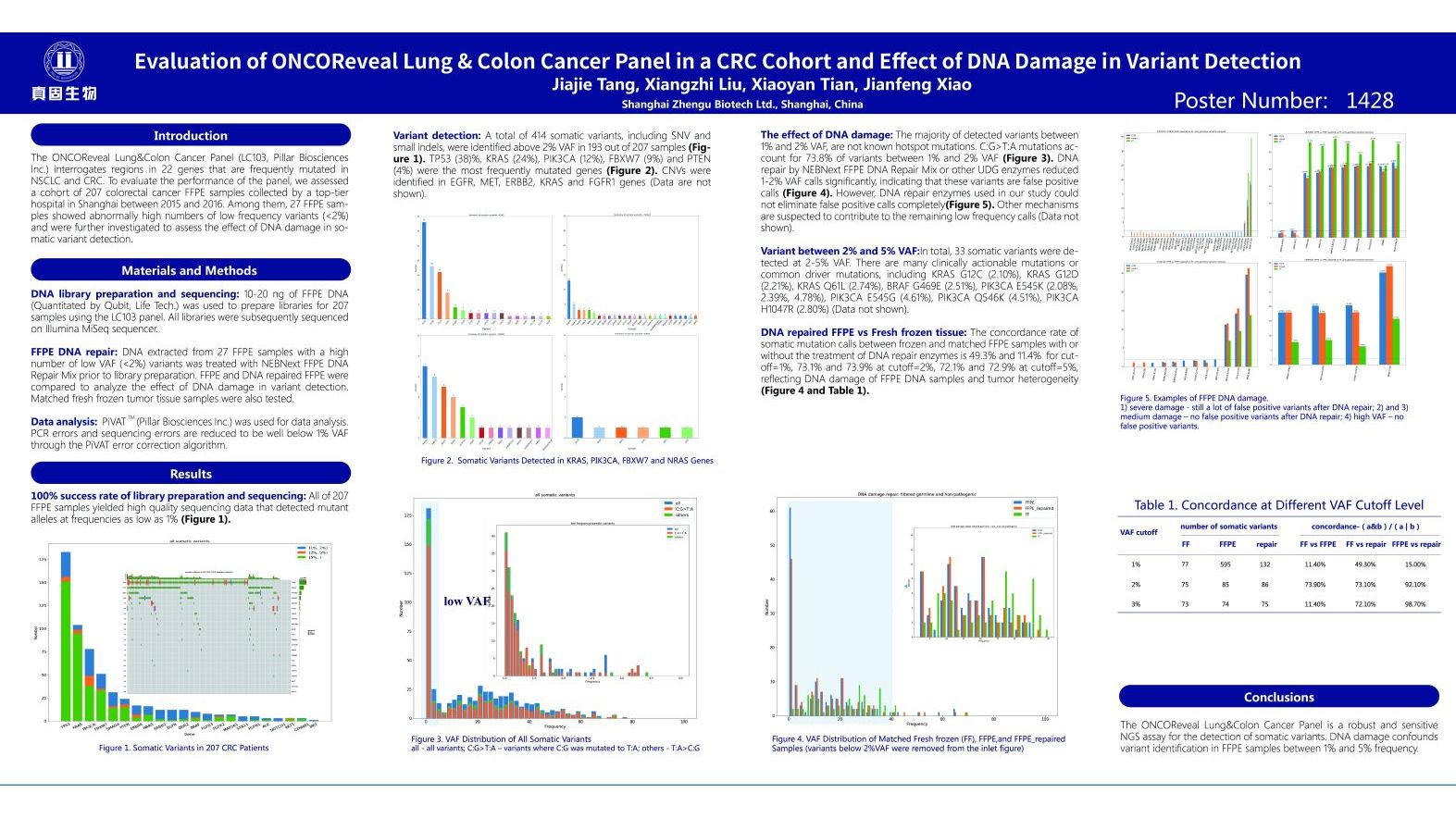
Evaluation of ONCOReveal Lung & Colon Cancer in a CRC Cohort and Effect of DNA Damage in Variant Detection
- Jiajie Tang, Xiangzhi liu, Xiaoyan Tian, Jianfeng Xiao
- AACR 2018
- Access a poster presented by Jianfeng Xiao, Ph.D. at the April 2018 American Association for Cancer Research (AACR) in Chicago Illinois. The poster presents results from over 200 FFPE samples used for evaluating the performance of the Pillar Biosciences’ ONCO/Reveal Lung & Colon Cancer Panel, with a subset of 27 samples selected for further analysis for DNA artifacts and effectiveness of repair.

Evaluation of the Pillar NGS SLIMap Cancer Hotspot Panel
- Francine B. de Abreu, Jason D. Peterson, Brian Dugan, Zhaohui Wang, Gregory J. Tsongails
- AMP 2017
- Access this poster presented by Fran de Abreu, Ph.D. at the recent Association for Molecular Pathology conference in Salt Lake City, UT November 2017. The poster presents results from a total of 15 FFPE samples used for evaluating the performance of the Pillar Biosciences’ ONCO/Reveal Solid Tumor Panel, with the amount of genomic DNA input ranging from 17 – 72 ng and a total of 24 samples run on an Illumina MiSeq system using v3 chemistry. A median on-target rate of 99.57% and a median mapping rate of 99.11% are reported, with 100% concordance of previously reported results.
Evaluation of the Pillar NGS SLIMamp Lung and Colon Hot Spots Panel
- Francine B. de Abreu, Jason D. Peterson, Brian Dugan, Zhaohui Wang, Gregory J. Tsongalis
- AMP 2016
- This poster was presented at the Association for Molecular Pathology 2016 conference held in Charlotte, NC and compared the performance of the Pillar Biosciences’ Lung and Colon Panel on an Illumina MiSeq™ to the Thermo Fisher Ampliseq™ Cancer Hotspot Panel v2 on the Ion Torrent PGM™. 15 previously-characterized FFPE samples found a high degree of concordance (90.0%, 27/30 variants), and the Pillar Biosciences’ assay was shown to be highly reproducible at 5 ng and 50 ng input DNA. Allele frequency measurement reproducibility was also evaluated. J. Peterson, F. Blumental de Abreu, Z. Wang, W. Wells, G. Tsongalis, “Evaluation of the Pillar NGS SLIMamp Lung and Colon Hot Spots Panel”. Dartmouth-Hitchcock Medical Center, Lebanon, NH; Pillar Biosciences, Natick, MA

